
Godzik Lab
BIMR
 | Target desc: NC_004718_SARS-human_Tor2_nsp2_805..2718_1914bp_638aa |
JCMM | JCSG Godzik Lab BIMR |
Crystallization
class: 2 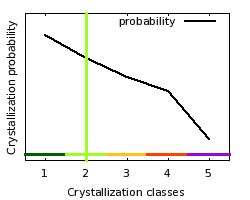 Crystallization classes 
| Comparison
of target features with distributions of crystallization probabilities
obtained from TargetDB 
| |||||||||||||||||||||||||
|
1...*...10....*...20....*...30....*...40....*...50....*...60....*...70....*...80....*...90....*..100 AVTRYVDNNFCGPDGYPLDCIKDFLARAGKSMCTLSEQLDYIESKRGVYCCRDHEHEIAWFTERSDKSYEHQTPFEIKSAKKFDTFKGECPKFVFPLNSK ....*..110....*..120....*..130....*..140....*..150....*..160....*..170....*..180....*..190....*..200 VKVIQPRVEKKKTEGFMGRIRSVYPVASPQECNNMHLSTLMKCNHCDEVSWQTCDFLKATCEHCGTENLVIEGPTTCGYLPTNAVVKMPCPACQDPEIGP ....*..210....*..220....*..230....*..240....*..250....*..260....*..270....*..280....*..290....*..300 EHSVADYHNHSNIETRLRKGGRTRCFGGCVFAYVGCYNKRAYWVPRASADIGSGHTGITGDNVETLNEDLLEILSRERVNINIVGDFHLNEEVAIILASF ....*..310....*..320....*..330....*..340....*..350....*..360....*..370....*..380....*..390....*..400 SASTSAFIDTIKSLDYKSFKTIVESCGNYKVTKGKPVKGAWNIGQQRSVLTPLCGFPSQAAGVIRSIFARTLDAANHSIPDLQRAAVTILDGISEQSLRL ....*..410....*..420....*..430....*..440....*..450....*..460....*..470....*..480....*..490....*..500 VDAMVYTSDLLTNSVIIMAYVTGGLVQQTSQWLSNLLGTTVEKLRPIFEWIEAKLSAGVEFLKDAWEILKFLITGVFDIVKGQIQVASDNIKDCVKCFID ....*..510....*..520....*..530....*..540....*..550....*..560....*..570....*..580....*..590....*..600 VVNKALEMCIDQVTIAGAKLRSLNLGEVFIAQSKGLYRQCIRGKEQLQLLMPLKAPKEVTFLEGDSHDTVLTSEEVVLKNGELEALETPVDSFTNGAIVG ....*..610....*..620....*..630....*... TPVCVNGLMLLEIKDKEQYCALSPGLLATNNVFRLKGG Legend
Download
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||