
Godzik Lab
BIMR
 | Target desc: NC_004718_SARS-human_Tor2_nsp8_12022..12615_594bp_198aa |
JCMM | JCSG Godzik Lab BIMR |
Crystallization
class: 2 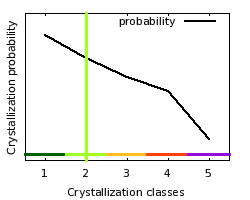 Crystallization classes 
| Comparison
of target features with distributions of crystallization probabilities
obtained from TargetDB 
| |||||||||||||||||||||||||
|
1...*...10....*...20....*...30....*...40....*...50....*...60....*...70....*...80....*...90....*..100 AIASEFSSLPSYAAYATAQEAYEQAVANGDSEVVLKKLKKSLNVAKSEFDRDAAMQRKLEKMADQAMTQMYKQARSEDKRAKVTSAMQTMLFTMLRKLDN ....*..110....*..120....*..130....*..140....*..150....*..160....*..170....*..180....*..190....*... DALNNIINNARDGCVPLNIIPLTTAAKLMVVVPDYGTYKNTCDGNTFTYASALWEIQQVVDADSKIVQLSEINMDNSPNLAWPLIVTALRANSAVKLQ Legend
Download
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||