
Godzik Lab
BIMR
 |
Target desc: NC_004718_SARS-human_Tor2_N_28120..29388_1269bp_422aa |
JCMM | JCSG Godzik Lab BIMR |
Crystallization
class: 5 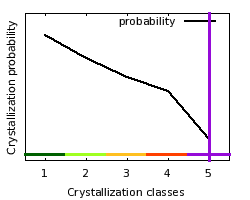 Crystallization classes 
| Comparison
of target features with distributions of crystallization probabilities
obtained from TargetDB 
| |||||||||||||||||||||||||
|
1...*...10....*...20....*...30....*...40....*...50....*...60....*...70....*...80....*...90....*..100 MSDNGPQSNQRSAPRITFGGPTDSTDNNQNGGRNGARPKQRRPQGLPNNTASWFTALTQHGKEELRFPRGQGVPINTNSGPDDQIGYYRRATRRVRGGDG ....*..110....*..120....*..130....*..140....*..150....*..160....*..170....*..180....*..190....*..200 KMKELSPRWYFYYLGTGPEASLPYGANKEGIVWVATEGALNTPKDHIGTRNPNNNAATVLQLPQGTTLPKGFYAEGSRGGSQASSRSSSRSRGNSRNSTP ....*..210....*..220....*..230....*..240....*..250....*..260....*..270....*..280....*..290....*..300 GSSRGNSPARMASGGGETALALLLLDRLNQLESKVSGKGQQQQGQTVTKKSAAEASKKPRQKRTATKQYNVTQAFGRRGPEQTQGNFGDQDLIRQGTDYK ....*..310....*..320....*..330....*..340....*..350....*..360....*..370....*..380....*..390....*..400 HWPQIAQFAPSASAFFGMSRIGMEVTPSGTWLTYHGAIKLDDKDPQFKDNVILLNKHIDAYKTFPPTEPKKDKKKKTDEAQPLPQRQKKQPTVTLLPAAD ....*..410....*..420.. MDDFSRQLQNSMSGASADSTQA Legend
Download
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||